This site contains downloads, links, and instructions for the ASHG 2019 Workshop: Interactive Introduction to Tools for Estimating Age-based Penetrance and Pathogenicity
Date/Time: Friday, October 18, 7:15-8:45 am
Contact Information
organizer: erosen[at]uw.edu
Please contact me with any questions or comments about the workshop and this website
analyze.myvariant: shirtsb[at]uw.edu
canrisk: aca20[at]medschl.cam.ac.uk
OpenMendel: jsinshei[at]ucla.edu
Slides from the workshop
OpenMendel: Slides for using the Julia command line
OpenMendel: Slides for using the Jupyter binder
See below for installing and using Julia
URLs
github.com/OpenMendel/GeneticCounseling_ASHG2019
File Downloads
We recommend that you use either Chrome or Firefox web browsers in order to get the downloads.
analyze.myvariant: example-pedigree-excel-file, example-penetrance-excel-file
BOADICEA: SNP-file1 (On a Windows machine, right click on the page that pops up, and choose ‘save as’. Your computer may assume that the file is a vCalender file, but this will not interfere with your ability to use the file. On a Mac, click the page with two fingers and choose ‘Save Page As’. If you are using Safari, click the page with two fingers and choose ‘Download linked file as…. ‘)
BOADICEA/CanRisk practical examples
OpenMendel: GeneticCounseling_ASHG2019. (On a Windows maching, this requires the use of unzipping software such 7Zip. Here are instructions to extract the files. An Apple machine should be able to automatically unzip the file.)
Background reading
analyze.myvariant: background, pedigree formatting, penetrance format
BOADICEA: PMID:30643217
OpenMendel: PMID:30915546
*IMPORTANT*
You will need to use the Julia programming language during part of the workshop. We highly recommend that you install and prepare Julia ahead of time, as this process can take some time:
Step 1: Download and install the Julia current stable release. To determine how many bits your Windows machine uses, follow the instructions given here.
Step 2: Start Julia (click on the icon). It will look something like this:
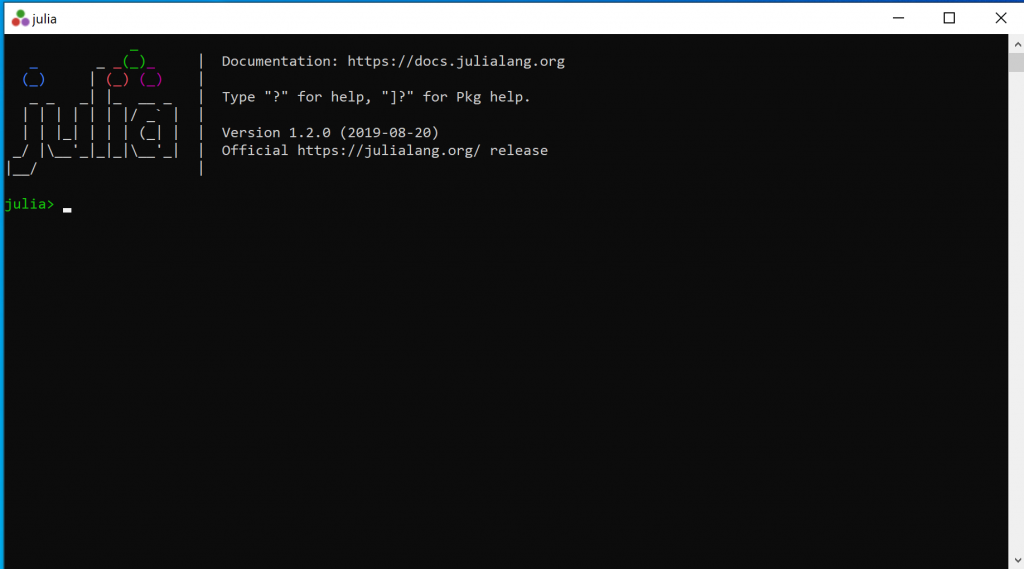
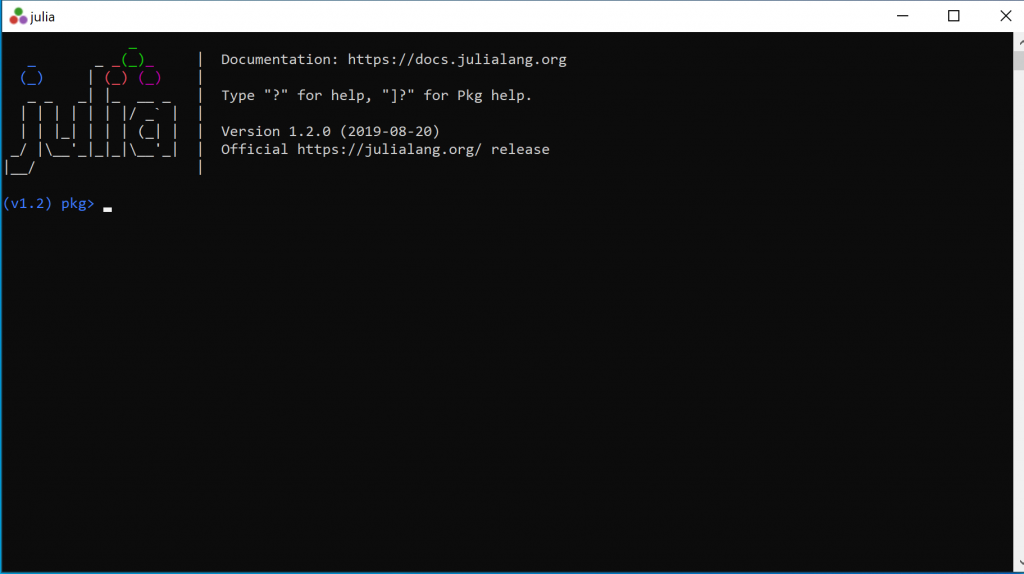
Step 3: Before you can do anything, you will need to install packages. You only need to install them once. To install the necessary packages, First, type
]It should now look something like this
Step 4: Copy and paste the following lines, one at a time. Be sure to wait for Julia to be done (i.e.,  reappears), before copying and pasting the next line. Note: Do not use ctrl-v to paste; instead use the mouse. Remember to hit enter after each line.
reappears), before copying and pasting the next line. Note: Do not use ctrl-v to paste; instead use the mouse. Remember to hit enter after each line.
add https://github.com/OpenMendel/SnpArrays.jl
add https://github.com/OpenMendel/MendelSearch.jl
add https://github.com/OpenMendel/MendelBase.jl
add https://github.com/OpenMendel/MendelGeneticCounseling.jl#ASHG2019
build SpecialFunctionsStep 6: Press the backspace key to exit the package mode
You have now set up Julia and can exit by typing ‘ctrl-d’. Below are a few more instructions that may prove helpful.
Navigating directories within Julia
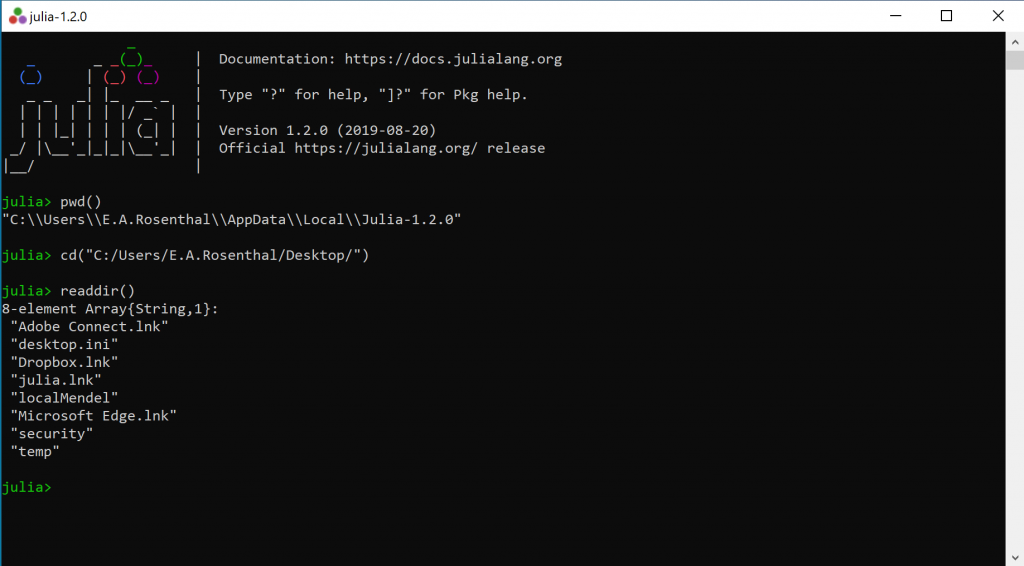
The Julia app is probably not in the same directory as the data you want to analyze. To see which directory you are in, type
pwd()To get to another directory, type the following, where <PATH> is the directory tree (see example below)
cd("C:/<PATH>/")To see what is in the directory, type
readdir()Here is an example using all three commands above
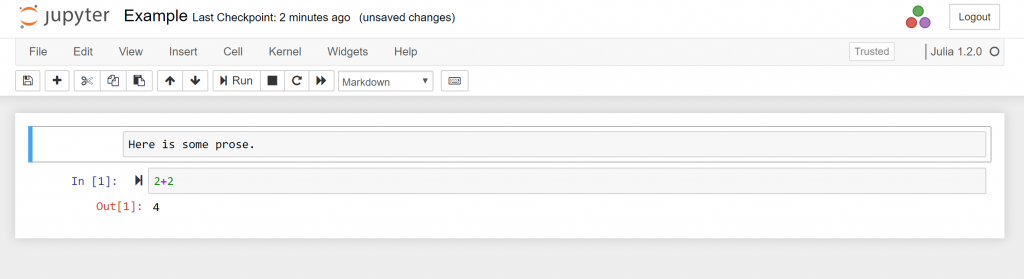
Using Jupyter notebooks with Julia
The term Jupyter derives from the coding languages Julia, Python and R. A Jupyter notebook is a simple way to combine prose, code and output from an analysis. It also makes it easier to navigate between directories. Below is a an example screenshot
To use Jupyter notebooks, you will need to install the package IJulia within the package environment (see steps 3 and 4 above)
]
add IJuliaPress the backspace key to exit the package environment, then type
using IJulia
notebook()Answer ‘y’, at the prompt. When the notebook() environment is ready, a web-page will open. You can navigate within that webpage to get to a current Jupyter (.ipynb) notebook in the GeneticCounseling_ASHG2019-master directory that you downloaded and unzipped earlier (under “File Downloads” at the top of this page).