Links
Selected Projects
Miscellaneous
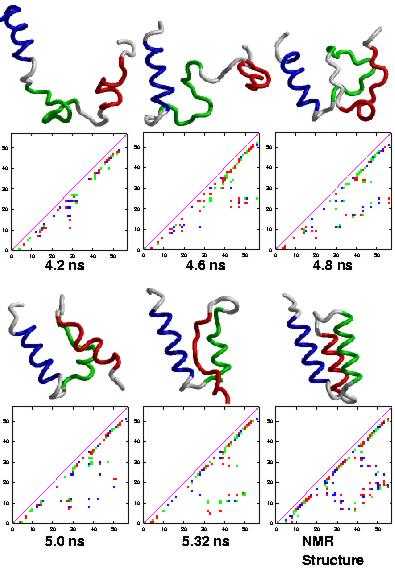
Figure Caption
The figure on the right shows mainchain traces and sidechain contact maps
for structures between 4.2 and 5.32 ns taken from a high temperature simulation
of the E-domain of Protein A. During this latter part of the simulation,
the protein re-formed native secondary structure and contacts, however
the topology of the helices mirrored that of the NMR structure:
H1 (red) is front in the simulation structures and in the rear in the NMR structure.
As expected at high temperature, the native-like state only existed transiently.
Times are given below the contacts maps, and the NMR structure is presented for comparison.
MPEG MOVIE (13 MB)
Quicktime MOVIE (83 MB)
Conformational Sampling in High Temperature Simulations