# Create map as before, but add polygons and labels before other elements.
g3 <- leaflet(data = wa_coverage) %>%
setView(-122.5487, 45.65366, zoom = 11) %>%
addControl(title, position = "topleft", className ="map-title") %>%
addTiles(group = "map-detail") %>%
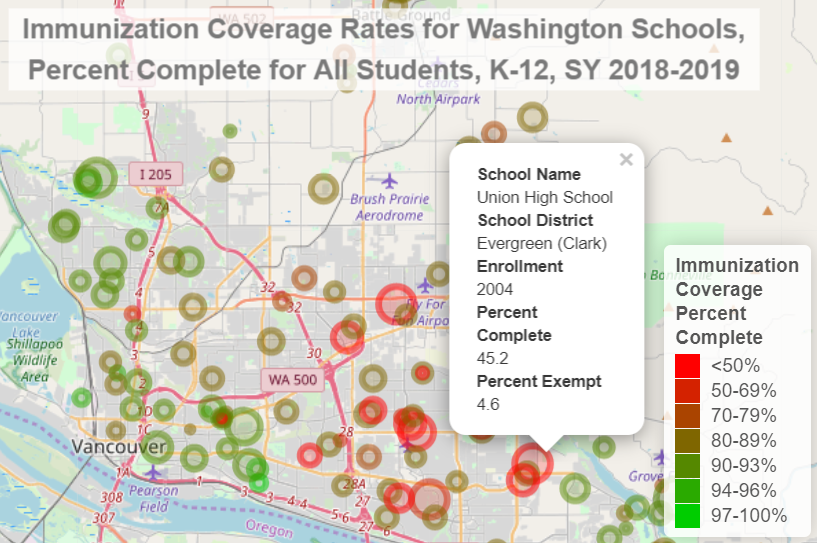
addLegend("bottomright", pal = pal, values = ~Pct,
title = "Immunization<br>Coverage<br>Percent<br>Complete",
opacity = 1, group = "legend") %>%
addPolygons(group = "districts", data = shapeData_clark, weight = 4,
color = 'gray', opacity = 0.4,stroke = TRUE, smoothFactor = 0.2,
fillOpacity = 0.3, fillColor = pal(shapeData_clark$Pct)) %>%
addLabelOnlyMarkers(group = "dist-labels", data = centers, lng = ~x, lat = ~y,
labelOptions = labelOptions(textsize='12px', opacity = 0.8, noHide = TRUE,
direction = 'top', textOnly = FALSE), label = htmlEscape(~NAME)) %>%
addCircleMarkers(group = "schools", lng = ~lon, lat = ~lat, color = ~pal(Pct),
radius = ~sqrt(Enrollment) * .3, popup = ~Popup, label = ~htmlEscape(label)) %>%
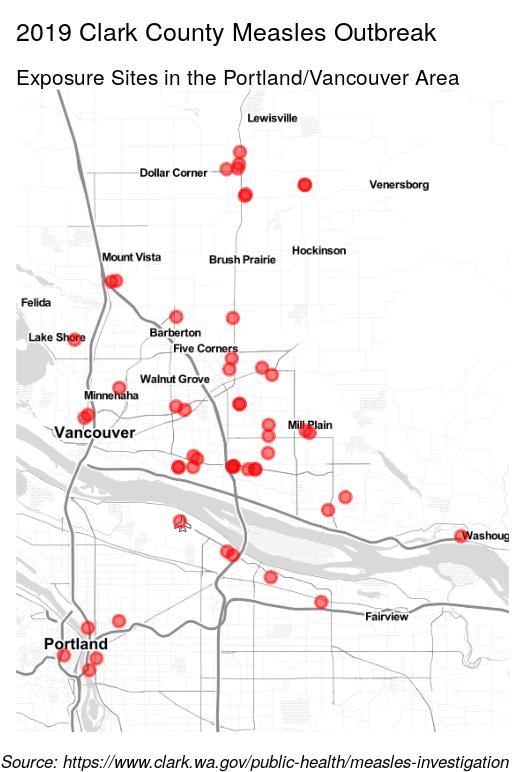
addMarkers(lng = ~lon, lat = ~lat, data = locations, group = "exposure",
icon = list(iconUrl = 'biohazard.svg', iconSize = c(20, 20)),
label = ~lapply(label, HTML), labelOptions = labelOptions(
style = list('background-color' = 'yellow'))) %>%
addLayersControl(baseGroups = c("map-detail", "no-map-detail"),
overlayGroups = c("legend", "districts", "dist-labels", "schools", "exposure"),
options = layersControlOptions(collapsed = TRUE))